| PAS fold | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
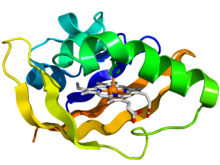 Crystallographic structure of the PAS domain of the bacterial oxygen sensor protein fixL.
[1] The protein is depicted as a rainbow colored cartoon (
N-terminus = blue,
C-terminus = red) while the
heme ligand is shown as sticks (carbon = white, nitrogen = blue, oxygen = red, iron = orange). | |||||||||||
| Identifiers | |||||||||||
| Symbol | PAS | ||||||||||
| Pfam | PF00989 | ||||||||||
| InterPro | IPR013767 | ||||||||||
| SMART | PAS | ||||||||||
| PROSITE | PDOC50112 | ||||||||||
| SCOP2 | 2phy / SCOPe / SUPFAM | ||||||||||
| CDD | cd00130 | ||||||||||
| |||||||||||
A Per-Arnt-Sim (PAS) domain is a protein domain found in all kingdoms of life. [2] Generally, the PAS domain acts as a molecular sensor, whereby small molecules and other proteins associate via binding of the PAS domain. [3] [4] [5] Due to this sensing capability, the PAS domain has been shown as the key structural motif involved in protein-protein interactions of the circadian clock, and it is also a common motif found in signaling proteins, where it functions as a signaling sensor. [6] [7]
Discovery
PAS domains are found in a large number of organisms from bacteria to mammals. The PAS domain was named after the three proteins in which it was first discovered: [8]
- Per – period circadian protein
- Arnt – aryl hydrocarbon receptor nuclear translocator protein
- Sim – single-minded protein
Since the initial discovery of the PAS domain, a large quantity of PAS domain binding sites have been discovered in bacteria and eukaryotes. A subset called PAS LOV proteins are responsive to oxygen, light and voltage. [9]
Structure
Although the PAS domain exhibits a degree of sequence variability, the three-dimensional structure of the PAS domain core is broadly conserved. [10] This core consists of a five-stranded antiparallel β-sheet and several α-helices. Structural changes, as a result of signaling, predominantly originate within the β-sheet. These signals propagate via the α-helices of the core to the covalently-attached effector domain. [11] In 1998, the PAS domain core architecture was first characterized in the structure of photoactive yellow protein (PYP) from Halorhodospira halophila. [10] In many proteins, a dimer of PAS domains is required, whereby one binds a ligand and the other mediates interactions with other proteins. [5]
Examples of PAS in organisms
The PAS domains that are known share less than 20% average pairwise sequence identity, meaning they are surprisingly dissimilar. [10] PAS domains are frequently found on proteins with other environmental sensing mechanisms. Also, many PAS domains are attached to photoreceptive cells. [12]
Bacteria
Often in the bacterial kingdom, PAS domains are positioned at the amino terminus of signaling proteins such as sensor histidine kinases, cyclic-di-GMP syntheses and hydrolases, and methyl-accepting chemotaxis proteins. [10]
Neurospora
In the presence of light, White Collar-1 (WC-1) and White Collar-2 (WC-2) dimerizes via mediation by the PAS domains, which activates translation of FRQ. [13]
Drosophila
In the presence of light, CLK and CYC attach via a PAS domain, activating the translation of PER, which then associates to Tim via the PER PAS domain. The following genes contain PAS binding domains: PER, Tim, CLK, CYC.
Arabidopsis
A PAS domain is found in the ZTL and NPH1 genes. These domains are very similar to the PAS domain found in the Neurospora circadian-associated protein WC-1. [14]
Mammals
The circadian clock that is currently understood for mammals begins when light activates BMAL1 and CLK to bind via their PAS domains. That activator complex regulates Per1, Per2, and Per3 which all have PAS domains that are used to bind to cryptochromes 1 and 2 ( CRY 1,2 family). The following mammalian genes contain PAS binding domains: Per1, Per2, Per3, Cry1, Cry2, Bmal, Clk, Pasd1.
Other mammalian PAS roles
Within Mammals, both PAS domains play important roles. PAS A is responsible for the protein-protein interactions with other PAS domain proteins, while PAS B has a more versatile role. It mediates interactions with chaperonins and other small molecules like dioxin, but PAS B domains in NPAS2, a homolog of the Drosophila clk gene, and the hypoxia inducible factor (HIF) also help to mediate ligand binding. [12] Furthermore, PAS domains containing the NPAS2 protein have been shown to be a substitute for the Clock gene in mutant mice who lack the Clock gene completely. [15]
The PAS domain also directly interacts with BHLH. It is typically located on the C-Terminus of the BHLH protein. PAS domains containing BHLH proteins form a BHLH-Pas protein, typically found and encoded in HIF, which require both the PAS domain and BHLH domain and the Clock gene. [16] [17] [18]
References
- ^ PDB: 1y28; Dunham CM, Dioum EM, Tuckerman JR, Gonzalez G, Scott WG, Gilles-Gonzalez MA (July 2003). "A distal arginine in oxygen-sensing heme-PAS domains is essential to ligand binding, signal transduction, and structure". Biochemistry. 42 (25): 7701–8. doi: 10.1021/bi0343370. PMID 12820879. S2CID 14090693.
- ^ Henry, Jonathan T.; Crosson, Sean (1 January 2011). "Ligand-binding PAS domains in a genomic, cellular, and structural context". Annual Review of Microbiology. 65: 261–286. doi: 10.1146/annurev-micro-121809-151631. PMC 3298442. PMID 21663441.
- ^ Liu, Yu C.; Machuca, Mayra A.; Beckham, Simone A.; Gunzburg, Menachem J.; Roujeinikova, Anna (1 October 2015). "Structural basis for amino-acid recognition and transmembrane signalling by tandem Per-Arnt-Sim (tandem PAS) chemoreceptor sensory domains". Acta Crystallographica Section D. 71 (10): 2127–2136. Bibcode: 2015AcCrD..71.2127L. doi: 10.1107/S139900471501384X. PMID 26457436.
- ^ Möglich, Andreas; Ayers, Rebecca A.; Moffat, Keith (14 October 2009). "Structure and signaling mechanism of Per-ARNT-Sim domains". Structure. 17 (10): 1282–1294. doi: 10.1016/j.str.2009.08.011. PMC 3092527. PMID 19836329.
- ^ a b Hennig, Sven; Strauss, Holger M.; Vanselow, Katja; Yildiz, Özkan; Schulze, Sabrina; Arens, Julia; Kramer, Achim; Wolf, Eva (28 April 2009). "Structural and Functional Analyses of PAS Domain Interactions of the Clock Proteins Drosophila PERIOD and Mouse PERIOD2". PLOS Biology. 7 (4): e1000094. doi: 10.1371/journal.pbio.1000094. PMC 2671562. PMID 19402751.
- ^ Ponting CP, Aravind L (November 1997). "PAS: a multi-functional domain family comes to light". Curr. Biol. 7 (11): R674–7. doi: 10.1016/S0960-9822(06)00352-6. PMID 9382818. S2CID 14105830.
- ^ Hefti MH, Françoijs KJ, de Vries SC, Dixon R, Vervoort J (March 2004). "The PAS fold. A redefinition of the PAS domain based upon structural prediction". Eur. J. Biochem. 271 (6): 1198–208. doi: 10.1111/j.1432-1033.2004.04023.x. PMID 15009198.
- ^ Möglich, Andreas; Ayers, Rebecca A.; Moffat, Keith (14 October 2009). "Structure and Signaling Mechanism of Per-ARNT-Sim Domains". Structure. 17 (10): 1282–1294. doi: 10.1016/j.str.2009.08.011. PMC 3092527. PMID 19836329.
- ^ Rosato, Ezio; Tauber, Eran; Kyriacou, Charalambos P. (1 January 2006). "Molecular genetics of the fruit-fly circadian clock". European Journal of Human Genetics. 14 (6): 729–738. doi: 10.1038/sj.ejhg.5201547. PMID 16721409.
- ^ a b c d Henry, Jonathan T.; Crosson, Sean (1 January 2011). "Ligand-Binding PAS Domains in a Genomic, Cellular, and Structural Context". Annual Review of Microbiology. 65: 261–286. doi: 10.1146/annurev-micro-121809-151631. PMC 3298442. PMID 21663441.
- ^ Möglich, A; Ayers, RA; Moffat, K (2009). "Structure and Signaling Mechanism of Per-ARNT-Sim Domains". Structure. 17 (10): 1282–94. doi: 10.1016/j.str.2009.08.011. PMC 3092527. PMID 19836329.
- ^ a b McIntosh, Brian; Hogenesch, John; Bradfield, Christopher (2010). "Mammalian Per-Arnt-Sim Proteins in Environmental Adaptation". Annual Review of Physiology. 72: 625–645. doi: 10.1146/annurev-physiol-021909-135922. PMID 20148691.
- ^ Harmer, Stacey L.; Panda, Satchidananda; Kay, Steve A. (28 November 2003). "Molecular Bases of Circadian Rhythms". Annual Review of Cell and Developmental Biology. 17: 215–253. doi: 10.1146/annurev.cellbio.17.1.215. PMID 11687489.
- ^ Somers, David; Schultz, Thomas; Kay, Steve; Milnamow, Maureen (2000). "ZEITLUPE Encodes a Novel Clock-Associated PAS Protein from Arabidopsis". Cell. 101 (3): 319–329. doi: 10.1016/S0092-8674(00)80841-7. PMID 10847686. S2CID 3013788.
- ^ Debruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM (May 2006). "A clock shock: mouse CLOCK is not required for circadian oscillator function". Neuron. 50 (3): 465–77. doi: 10.1016/j.neuron.2006.03.041. PMID 16675400. S2CID 19028601.
- ^ Jones, Susan (1 January 2004). "An overview of the basic helix-loop-helix proteins". Genome Biology. 5 (6): 226. doi: 10.1186/gb-2004-5-6-226. PMC 463060. PMID 15186484.
- ^ Ke, Qingdong; Costa, Max (1 November 2006). "Hypoxia-Inducible Factor-1 (HIF-1)". Molecular Pharmacology. 70 (5): 1469–1480. doi: 10.1124/mol.106.027029. PMID 16887934. S2CID 2522614.
- ^ Wang, G. L.; Jiang, B. H.; Rue, E. A.; Semenza, G. L. (6 June 1995). "Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension". Proceedings of the National Academy of Sciences of the United States of America. 92 (12): 5510–5514. Bibcode: 1995PNAS...92.5510W. doi: 10.1073/pnas.92.12.5510. PMC 41725. PMID 7539918.